Mentioned project were developed in my free time and/or with cooperation with my students. Some of them are long-term and still open and ready for further improvements.
Virtual Table
Real-time Simulations, Parallel Computing, and Visualization
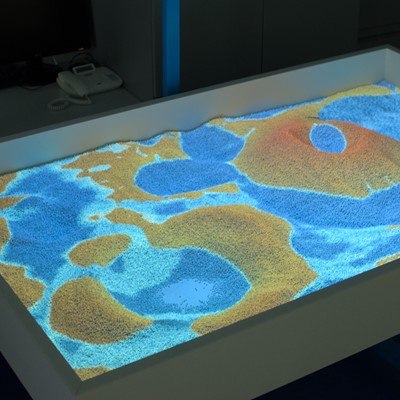
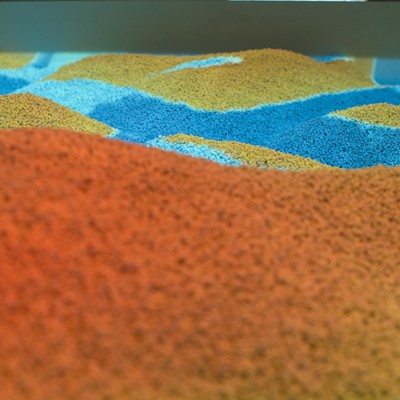
This project illustrates a close connection between three research areas - simulation, computing, and visualization. Real-time simulations represent advanced tasks in the area of programming because of the higher requirements to computations, hardware, programming skills and knowledge. Parallel computing is represented by software components that use NVIDIA graphics cards and CUDA. Finally, visualization was implemented in OpenGL. Rain, water flow, infiltration and evaporation were implemented. User interaction was implemented with the use of Microsoft Kinect v2. Color and depth images were pre-processed in CUDA. Terrain triangulation and some simulation parts were done in CUDA. The rest of simulation was computed in GLSL just to increase the performance of following visualization in OpenGL.
Current state of the application:
- Hardware implementation of the virtual table
- Visualization toolkit
- Water simulation
Co-authors:
- Ing. Michal Radecký, Ph.D.
- Ing. Jan Kuba
Bio-inspired Methods
General Toolkit for Large Scale Data Analysis
Processing large datasets generated by research or industry represents one of the most important challenges in data mining. Ever-growing datasets present an opportunity for discovering previously unobserved patterns. Although analyzing data is the major issue of data mining, general processing of large data can be extremely time consuming. Bio-inspired computations provide a set of powerful methods and techniques based on the principles of biological, natural systems, e.g. evolutionary algorithms, ant colony, neural networks, swarm intelligence, etc. that complement the traditional techniques of data mining and can be applied in places where the earlier approaches have encountered difficulties. Bio-inspired methods have been successfully applied in various research areas from software engineering and data processing to chemical engineering and molecular biology. The main goal of this project consists in the implementation of powerful bio-inspired methods and algorithms for data mining that will reflect current trends in data processing and hardware evolution.
Current state of the application:
- Fast GPU implementations of selected bio-inspired methods
- Visualization toolkit
Co-authors:
- Ing. Jan Janoušek
Signal processing
EEG Signal Data Processing
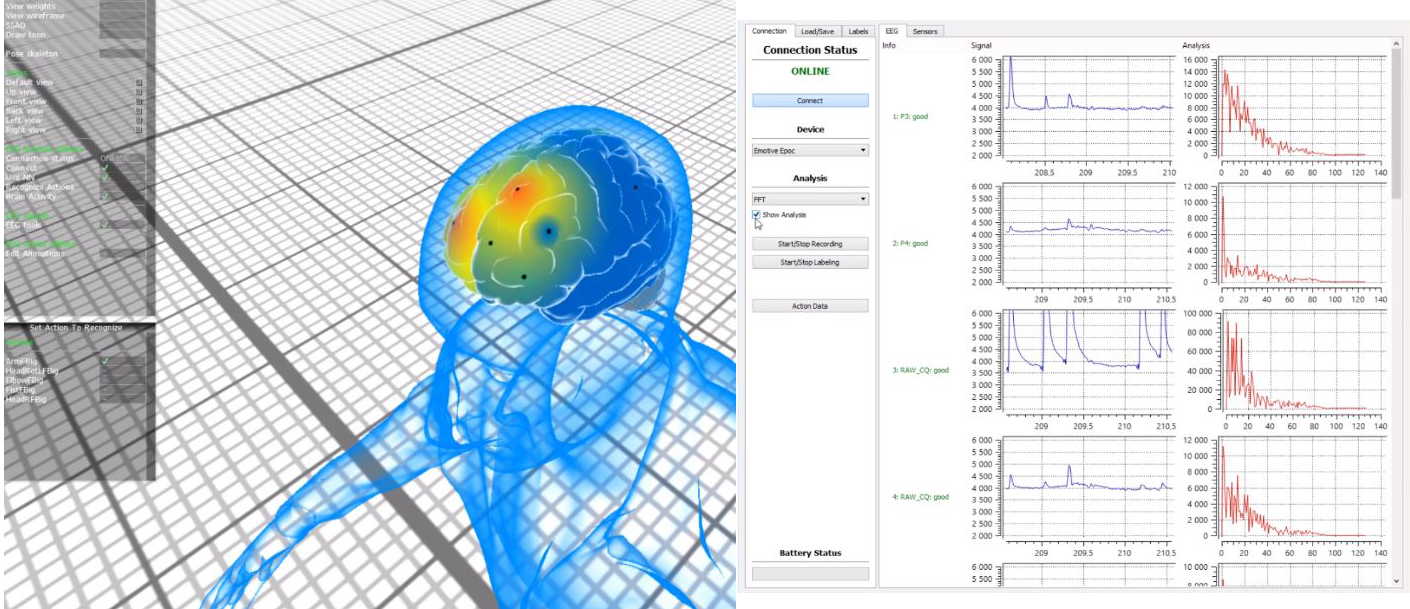
Human-Computer interaction is a very important issue that has been addressed in many ways. The Brain Computer Interface (BCI) is an attempt to communicate directly a brain with a computer. This is of the utmost interest for people with severe motor disabilities, who cannot use the standard communication devices like keyboards or touchpads. Most usually, the BCI relies on non-invasive EEG (electroencephalogram) electrodes, which are attached to the scalp. The electrodes detect the EEG signals related to motor intentions, like the preparation to move the left hand, or just imagining making such movement. Once the EEG signal has been decoded, it can be used to move a cursor on the screen, or to execute commands in a computer. For instance, the intention to move the right hand can be used to move the cursor to the right, and so on.
Decoding the EEG signal is not a straightforward task. The signal is very weak and many artifacts can be present (just blinking an eye may add noise to the signal). But most importantly, there is no simple function to map EEG signals to intentions. In addition, the mapping function can change from person to person, or even for the same person on different days. A common approach is to inductively learn the mapping function from hundreds of labelled data. For this task, inductive algorithms like Neural Networks or Support Vector Machines, can be used.
The current state of the application:
- EEG EPOC headset utilization
- C++, Qt application that covers basic funcionality (EPOC EDK included)
- Basic concept of GPU library for data manipulation
Co-authors:
- Ing. Martin Barteček
- Ing. Jakub Rodzenák
DNA Analysis
The sequence of the human genome is of interest in several respects. It is the largest genome to be extensively sequenced so far, being 25 times as large as any previously sequenced genome and eight times as large as the sum of all such genomes. Much work remains to be done to produce a complete finished sequence, but the vast trove of information that has become available through this collaborative effort allows a global perspective on the human genome. The genomic landscape shows marked variation in the distribution of a number of features, including genes, transposable elements, recombination rate, etc. There appear to be about 30,000–40,000 protein-coding genes in the human genome. However, the genes are more complex, with more alternative splicing generating a larger number of protein products.
We focused on applications of several data-mining and bio-inspired methods in the area of human DNA analysis. From the practical point of view, an effective and precise analysis and subsequent classification can play an important role in medical care. That is why we have established a close cooperation with the Department of Immunology, FM Palacky University & University Hospital Olomouc.
The current state of the application:
- Desktop application for data analysis and visualization
- Utilization of R statistics
- Implementation of the Formal Concept Analysis
Co-author:
- Ing. Pavel Dohnálek
Protein Analysis
The protein data bank (PDB) is a database containing information on experimentally determined three-dimensional structures of proteins, nucleic acids, and complex assemblies. In recognition of the growing international and interdisciplinary nature of structural biology, three organizations have formed a collaboration to oversee the newly formed worldwide Protein Data Bank (wwPDB; http://www.wwpdb.org/). The Research Collaboratory for Structural Bioinformatics (RCSB), the Macromolecular Structure Database (MSD) at the European Bioinformatics Institute (EBI) and the Protein Data Bank Japan (PDBj) at the Institute for Protein Research in Osaka University will serve as custodians of the wwPDB, with the goal of maintaining a single archive of macromolecular structural data that is freely and publicly available to the global community. The Protein Data Bank (PDB) is a repository for 3-D structural data of proteins and nucleic acids. This data, typically obtained by X-ray crystallography or NMR spectroscopy, is submitted by biologists and biochemists from around the world. We want to use new method with semantic data mining to denote a protein sequence for researching the protein classification and structure prediction problem. Those vectors in PDB files are used to classify proteins combined with the machine learning technology, such as support vector machine (SVM). We will do experiments of family-level protein classification on structural classification of PDB database to show the performance and hope this method is better than that of the others. Biological databases are the semi structured data. The search procedure in the semi structured data is hard. A new system will be developed to make the search and the comparison of the protein data banks easy and it will give the data in a structured way. Since the meta-data is created, the data can be easily found by the users. Biological data in PDB has its own challenging characteristics, which are huge data, heterogeneous distributed Data, and frequently updated data. Overcoming data heterogeneity has become vital by adding semantics, and defining common ontology, where efficient data integration techniques are required.
Current state of the applications
- Protein structure reader with support of PDB files.
- C++, Qt application for visualization and data analysis
- CUDA based libraries for pattern matching
FEM Visualization Toolbox
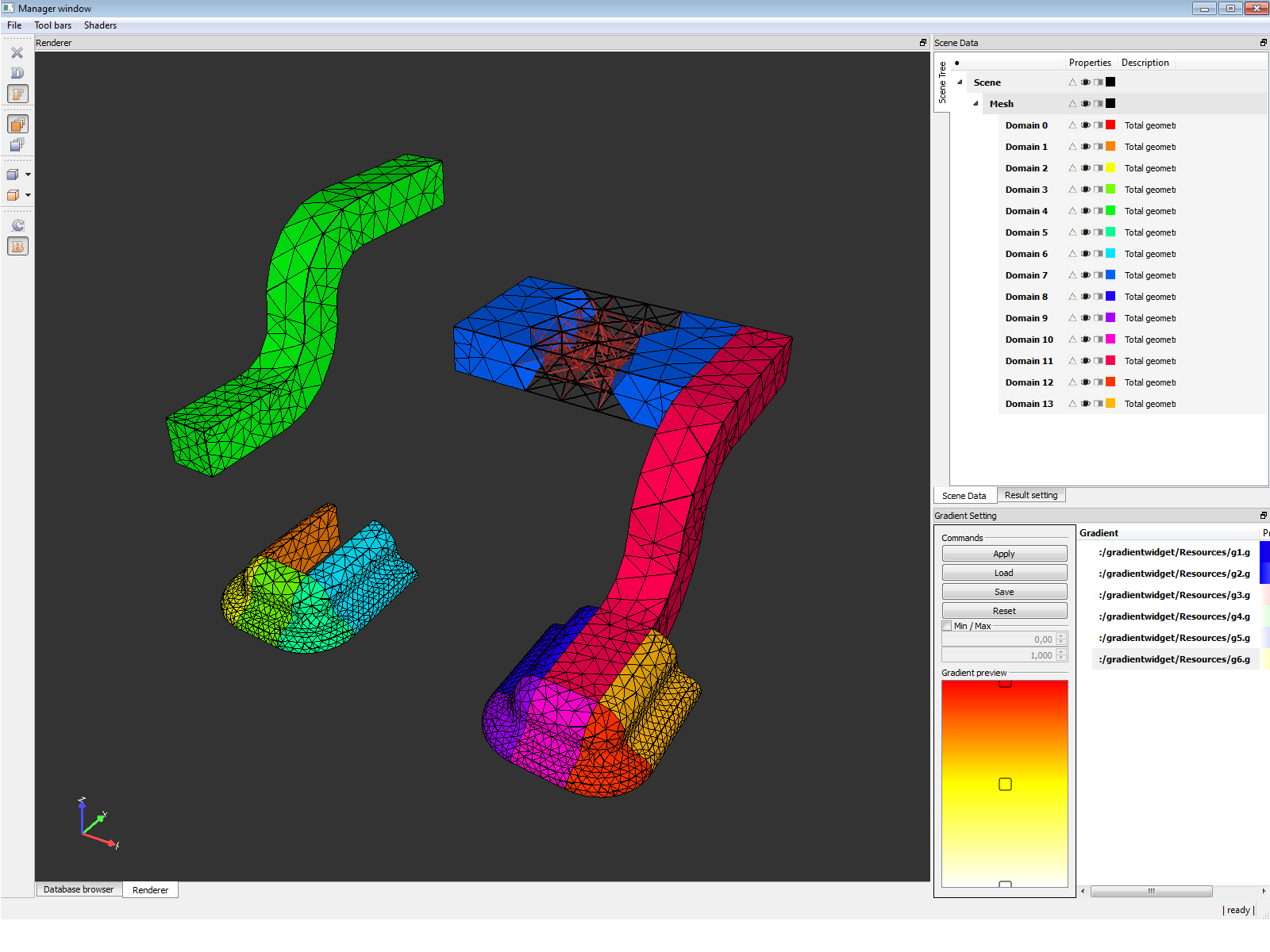
Finite element analysis (FEA) has become commonplace in recent years, and is now the basis of a multibillion dollar per year industry. Numerical solutions to even very complicated stress problems can now be obtained routinely using FEA, and the method is so important that even introductory treatments of Mechanics of Materials should outline its principal features. We focused on the data structures and visualization which will be suitable for Finite Element Analysis with great accept on processing of large models.
Current state of the applications
- C++, Qt application for visualization of Finite Element Models and results
- New data structures and database schema
- Data manipulation with respect to real-time rendering